Abstract
Introduction: The recombination of chromosomes 4 and 14 (t(4;14)) is a primary, predominantly clonal event in newly diagnosed multiple myeloma (ndMM) that is present in ~15% of patients. The translocation results in enhancer regions from the immunoglobulin heavy chain locus upregulating the expression of NSD2 and FGFR3 genes implicated in the disease biology of this subset of MM patients (Chesi et al. Blood. 1998, Keats et al, Leuk Lymph. 2006). The presence of t(4;14) translocation is a considered a biomarker of aggressive disease and is part of the Revised International Staging System (R-ISS) for clinical risk stratification. However, historically only ~40% of t(4;14) patients are high-risk based on the GEP70 gene expression signature. (Weinhold et al. Leukemia. 2016) Our previous analysis of a large cohort of ndMM patients described the genomic features of t(4;14) vs ndMM overall population demonstrating that only ~25% of t(4;14) patients died within 24 months of diagnosis and described biomarkers in this high-risk subset. This analysis identified both known and novel aberrations in ndMM, including some that were associated with high-risk t(4;14) (Ortiz et al Blood. 2019; 134 (Suppl_1):366). In this updated analysis, we provide a more robust analysis of the t(4;14) dataset and demonstrate the prognostic value of the NSD2 breakpoint location.
Methods: We generated a large genomic dataset from t(4;14) ndMM patients with whole genome sequencing (WGS) and RNA-seq from a TOUL dataset (t(4;14) N=114) patients treated in routine practice), the IFM2009 trial (N=19), and the Myeloma Genome Project (MGP) (N=34) for discovery and validation. Gene expression, copy number aberration, single nucleotide variant and translocations were derived from RNAseq and WGS profiling of biopsies from patients aged less than 75 years who received transplant, and integrated with clinical information (including age, OS). Cytogenetic assessments from WGS were made by MANTA and used to identify translocation DNA breakpoint location.
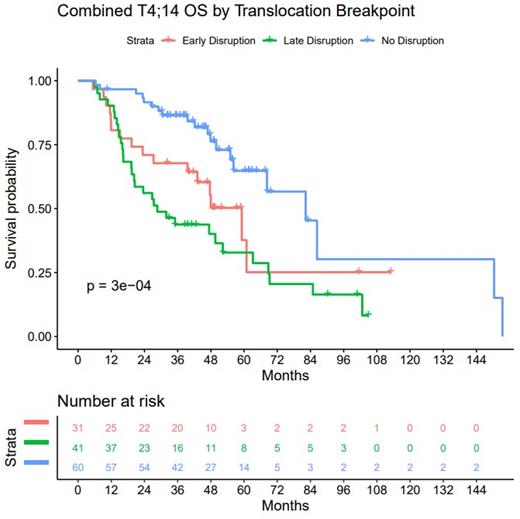
Results: In all datasets, three DNA breakpoint locations were identified, and based on their position with respect to the NSD2 gene named "no-disruption" (upstream of NSD2 gene), "early-disruption" (in the 5' UTR of NSD2 gene) and "late-disruption" (in the coding region of NSD2 gene). Using paired RNA-seq data, we identified IGH-NSD2 RNA fusion transcripts relative to the breakpoints that corresponded with previously described NSD2 isoforms. "No-disruption" and "early-disruption" breakpoints predominantly produced a fusion transcript (MB4-1) that retained the full coding sequence of the gene, while the "late-disruption" produced truncated fusion transcripts (MB4-2/3). We conducted survival analysis in our datasets based on both DNA breakpoint location and RNA fusion transcripts. This analysis demonstrated a significant difference in outcome between the patient samples with "no-disruption" and the "late-disruption" breakpoints that associated with good and poor OS, respectively (OS pval < 3e-4) in the discovery TOUL dataset. Patients with "late-disruption" had a median OS of 28.64 mo vs 59.18 mo for "early disruption" and 82.26 mo for those with "no disruption" (Figure). This association was replicated in an independent dataset (MGP N=33, replication pval<4.3e-5). The mOS difference of patients based on which fusion transcript they express is less than the difference based on breakpoint (mOS MB4-1 = 47.38 mo. vs. MB4-2/3 = 60.89 mo.). These analyses demonstrate that the breakpoint location has a stronger association with outcome than fusion transcript expression.
Conclusion: From a large genomic dataset, we were able to discover and validate a clear association between the translocation breakpoints and survival outcome in t(4:14) ndMM patients. While prospective validation is needed before clinical application of our finding, molecular identification of high-risk t(4;14) patients using DNA breakpoint location may enable proper risk classification for this patient group at diagnosis, and would provide improved opportunities for risk-adjusted therapy and identification of a therapeutic target for this high-risk subpopulation. Ongoing work on mutations, copy number, and differential gene expression analyses between translocation breakpoint sub-groups and will be presented.
Stong: Bristol Myers Squibb: Current Employment, Current equity holder in publicly-traded company. Ortiz: Bristol Myers Squibb: Current Employment. Towfic: Bristol Myers Squibb: Current Employment, Current equity holder in publicly-traded company. Pierceall: Bristol Myers Squibb: Current Employment, Current equity holder in publicly-traded company. Flynt: Bristol Myers Squibb: Current Employment. Thakurta: Bristol Myers Squibb: Current Employment, Current equity holder in publicly-traded company, Patents & Royalties.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal